Fine-tuning Pre-trained Deep Learning Models
Introduction
The enduring struggle against plant diseases and pests constitutes a significant issue in the realm of agriculture, with grave ramifications for global food security. This challenge becomes more prominent in horticulture, where the accurate diagnosis and timely identification of these ailments are critical to effective integrated pest management.
Traditional diagnostic methods, primarily based on visual inspection by experts, have limitations. They can be time-consuming and prone to inaccuracies. But, the landscape of diagnostics is rapidly changing, driven by the disruptive power of artificial intelligence (AI), more specifically, deep learning. By leveraging complex algorithms that learn from and mimic human intelligence, deep learning is reshaping many fields, including the way we detect and manage plant diseases.
This blog post will outline my recent project where I applied deep learning techniques to classify plant diseases. The experience allowed me to hone my skills, solve complex problems, and contribute to a field with far-reaching societal implications. I was given the priviledge of presenting this project at the Association of Horticulture Science Conference, in Orlando, FL 2024, to horticultural professionals and researchers.
The Project
At the heart of this project, I worked with three well-regarded deep learning models: ResNet50, InceptionV3, and InceptionResNetV2. These models represent some of the cutting-edge developments in the field of AI, each with its unique architecture designed to process data in a manner that simulates the human brain’s neural network.
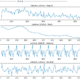
The models were trained and tested on the PlantVillage dataset, comprising over 20,000 images of both healthy and disease-affected leaves from potatoes, bell peppers, and tomatoes. The dataset was subject to data augmentation techniques, such as rotation, flipping, and zooming. The goal was to enhance the models’ robustness and prevent overfitting, a common issue where a model learns the training data too well and performs poorly on unseen data.
Following this, I fine-tuned the models. This involved adjusting their architecture by adding new layers and tweaking hyperparameters to better suit our specific task of plant disease detection. This fine-tuning process was informed by previous studies and a fair bit of trial and error.
To optimize the learning process and mitigate the loss, I used the Adam optimizer and categorical cross-entropy as the loss function. To further prevent overfitting and enhance the models’ ability to generalize, dropout layers were incorporated into the architecture. These layers randomly ignore neurons during training, reducing the model’s dependency on any single neuron and promoting a more holistic learning process.
Skills Acquired & Lessons Learned
This project was an invaluable learning journey, during which I acquired and sharpened several vital skills.
- Deep Learning: The practical experience with ResNet50, InceptionV3, and InceptionResNetV2 models allowed me to deepen my understanding of these algorithms’ underlying mechanics.
- Data Augmentation: Working with a sizable dataset allowed me to explore various data augmentation techniques. This experience offered insights into the ways of improving model performance and balancing the data.
- Model Fine-Tuning: Fine-tuning the pre-trained deep learning models was a key skill I developed. This involved experimenting with different architectures and hyperparameters, thus honing my analytical and problem-solving abilities.
- Analyzing Model Performance: Through evaluating the models using precision, recall, and F1-score, I gained a deeper understanding of these metrics and their importance in assessing a model’s performance.
Reflecting on the project, the primary challenge was managing imbalanced datasets, which led to models with high accuracy but low performance in other metrics. Moving forward, I aim to explore further techniques to handle such datasets, thereby improving the models’ classification capabilities.
Conclusion
The potential of deep learning in revolutionizing plant disease detection and, by extension, bolstering global food security, is immense. While the journey has its challenges, the results from this project underscore the value that such technologies can bring to the field.
I look forward to future projects where I can continue to refine these models, balance the dataset for individual classes, and fine-tune the hyperparameters. By doing so, we can move closer to making deep learning an indispensable tool in plant disease detection, driving forward our fight for global food security.


 WildRift Match Data Analysis and Dashboard
WildRift Match Data Analysis and Dashboard